Science
Advanced Simulations Reveal RNA Folding Breakthroughs
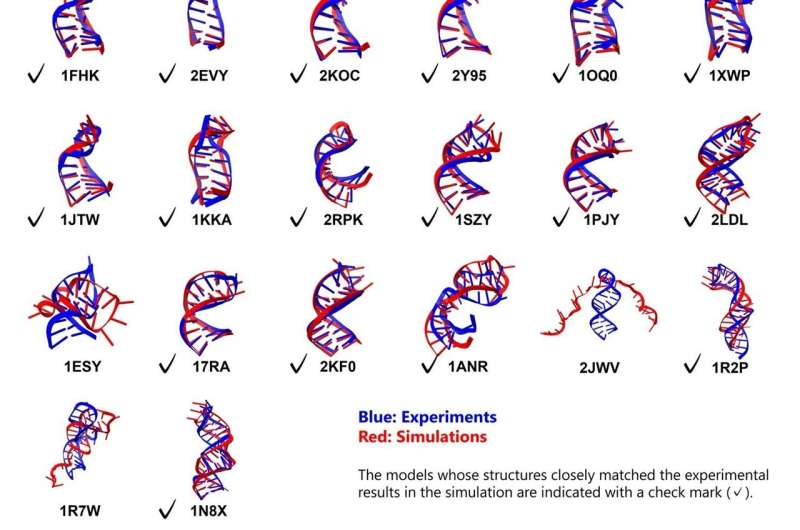
Researchers at the Tokyo University of Science have successfully advanced the understanding of RNA folding through innovative molecular dynamics simulations. In a study published on November 4, 2025, Associate Professor Tadashi Ando evaluated modern computational methods to predict RNA structures with unprecedented accuracy. This breakthrough is particularly timely as the global demand for RNA-based therapeutics continues to grow.
Understanding RNA’s Complex Structures
Ribonucleic acid (RNA) plays a crucial role in various biological processes, extending its functions beyond merely acting as a messenger for genetic information. RNA is integral in gene regulation, processing, and maintenance across all life forms. Its versatility is linked to its ability to fold into complex three-dimensional shapes, known as secondary and tertiary structures. Understanding these structures is essential for harnessing RNA’s potential in biotechnology.
Historically, simulating RNA folding from an initially unfolded state has posed significant challenges. Many previous molecular dynamics (MD) studies achieved accurate results only for simple stem-loop RNA structures. This limitation has underscored the need for improved computational models that can handle the complexities of larger and more diverse RNA libraries.
Innovative Approach to RNA Folding
In his recent research, Dr. Ando utilized a combination of two advanced computational tools: the DESRES-RNA atomistic force field and the GB-neck2 implicit solvent model. This approach approximates the surrounding fluid as a continuous medium, which significantly accelerates the sampling of conformations compared to traditional explicit solvent models that require more computational resources.
The study evaluated 26 RNA stem-loops, with lengths ranging from 10 to 36 nucleotides, incorporating various structural features such as bulges and internal loops. All simulations commenced from fully extended, unfolded conformations, allowing for a comprehensive analysis of the folding process.
The results revealed a high degree of stability and accuracy in the folding predictions. Notably, 23 out of the 26 RNA molecules folded into their expected structures, demonstrating the effectiveness of the combined modeling tools. For the simpler stem-loops, the simulations achieved remarkable accuracy, with root mean square deviation (RMSD) values of less than 2 Å for the stems and less than 5 Å for the overall molecules.
Among the more complex motifs, five out of eight with bulges or internal loops were accurately folded. The simulations even uncovered distinct folding pathways for these challenging structures. “Previous studies have reported only two or three folding examples of simple stem-loop structures of approximately 10 residues, whereas this study deals with structures of much greater scale and complexity,” Dr. Ando noted.
This successful simulation of diverse RNA structures marks a significant milestone in computational biology. It validates the chosen combination of force field and solvent models as a reliable foundation for exploring large-scale conformational changes in RNA.
While the stem regions showed high accuracy, the loop regions were less precise, indicating that the implicit solvent model parameters require further optimization. This is particularly true for modeling non-canonical base pairs and understanding the critical roles of divalent cations, such as magnesium.
The implications of this research extend to RNA-based medical applications. Accurate predictions of RNA folding are vital for designing targeted drugs, which could lead to new treatments for genetic disorders, viral infections like COVID-19 and influenza, and certain cancers.
Dr. Ando concluded, “The ability to reproduce the overall folding of the basic stem-loop structure is an important milestone in understanding and predicting the structure, dynamics, and function of RNA using highly accurate and reliable computational models. I expect this computational technique to lead to applications in RNA molecule design and drug discovery.”
This groundbreaking research not only enhances our understanding of RNA folding but also sets the stage for future developments in biotechnology and medicine.
-

 Science4 weeks ago
Science4 weeks agoUniversity of Hawaiʻi Joins $25.6M AI Initiative to Monitor Disasters
-

 Lifestyle2 months ago
Lifestyle2 months agoToledo City League Announces Hall of Fame Inductees for 2024
-

 Business2 months ago
Business2 months agoDOJ Seizes $15 Billion in Bitcoin from Major Crypto Fraud Network
-

 Top Stories2 months ago
Top Stories2 months agoSharp Launches Five New Aquos QLED 4K Ultra HD Smart TVs
-

 Sports2 months ago
Sports2 months agoCeltics Coach Joe Mazzulla Dominates Local Media in Scrimmage
-

 Politics2 months ago
Politics2 months agoMutual Advisors LLC Increases Stake in SPDR Portfolio ETF
-

 Health2 months ago
Health2 months agoCommunity Unites for 7th Annual Walk to Raise Mental Health Awareness
-

 Science2 months ago
Science2 months agoWestern Executives Confront Harsh Realities of China’s Manufacturing Edge
-

 World2 months ago
World2 months agoINK Entertainment Launches Exclusive Sofia Pop-Up at Virgin Hotels
-

 Politics2 months ago
Politics2 months agoMajor Networks Reject Pentagon’s New Reporting Guidelines
-

 Science1 month ago
Science1 month agoAstronomers Discover Twin Cosmic Rings Dwarfing Galaxies
-

 Top Stories1 month ago
Top Stories1 month agoRandi Mahomes Launches Game Day Clothing Line with Chiefs







